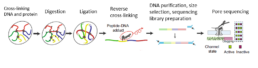
Pore-C
PORE-C is a modification of the Hi-C protocol, whereby short-read sequencing is replaced with Nanopore long-read sequencing methods.
High-order 3D interactions between more than two genomic loci are common in human chromatin, but their role in gene regulation is unclear. Previous high-order 3D chromatin assays either measure distant interactions across the genome or proximal interactions at selected targets. To address this gap, Pore-C was developed, which combines chromatin conformation capture (3C) with nanopore long-read sequencing of concatemers to profile proximal high-order chromatin contacts at the genome scale [1].
A few methods that generate multiway chromatin contacts have been developed, including genome architecture mapping (GAM), ChIA-drop, split-pool recognition of interactions by tag extension (SPRITE), Tri-C, multicontact 4C, concatemer ligation assay (COLA), and Pore-C [2].
Pore-C stands out because it can capture global high order multiway contacts, is technically simple, and captures DNA methylation simultaneously in a cell population. Because multiway contacts reflect synergistic chromatin interactions rather than multiplemutually exclusive interactions of different alleles, we can use Pore-C to reveal single-allele topology within designated genomic regions in populations of cells [2].
Plateformes à contacter pour ce domaine d’expertise