“Long Read” Sequencing
Based on SMRT technology (single molecule real time technology), RSII and Sequel sequencing enables producing very long readings to solve the coverage of low complexity regions and determine structural variants and haplotypes.
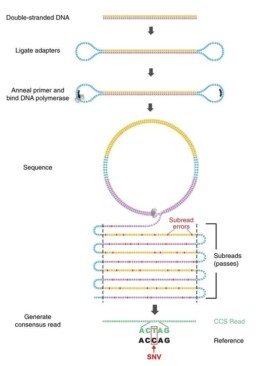
A SMRT library is built from fragments of 10 to 175kb. Once the ends have been repaired, single-stranded adapters are attached to them creating a loop (SMRT bell), anchoring point for DNA polymerase (Figure 1).
A complex comprising insert , adapter and polymerase is loaded, immobilised in each well of the sequencing SMRT cells and illuminated by a Zero Mode Waveguides (ZMW). This system detects fluorescent signals in real time during polymerisation (Figure 2).
The sequencing method has evolved with the HiFi version (Wenger AM et al. Nat. Biotech. 2019). Sequencing of the circularised insert is no longer obtained by a single pass of the polymerase (CLR) but by successive ‘ rounds’. The CLR (Continuous Long Read) mode enables inserts from 25 to 175kb to be analysed: multiple fragments of the same region will have to be sequenced to determine a consensus sequence with 90% acuracy. Using HiFi mode, replicate reading of the insert will produce a high-quality (99%) consensus sequence (CCS: Circular Conscensus Sequence) from a single 10-20kb fragment.