Targeted spatial omics methods using RNA probes
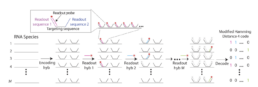
These methods are appropriate when there are specific molecular entities of interest to identify cellular states, identity, and function. RNA probes and antibodies, such as endogenous transcripts or proteins, are the most ubiquitous detection methods. The mRNA probes (specific for sequences of targeted transcripts) can be used to design a panel for targeted spatial profiling. Different multiplexed RNA FISH (fluorescence in situ hybridization) methods use slightly different approaches to probe design, imaging, and stripping [1]. Specifically, high multiplexing is achieved by using spectral barcoding (specific combination of fluorophores each targeting segments of an RNA resolved by microscopy) and temporal barcoding (multiple rounds of probe hybridization and stripping to create predefined color sequence). Recently, the combination of two barcoding methods and more complicated barcoding strategies can increase the number of molecular entities that can be profiled. To overcome potential errors and dropouts, most probe-based methods utilize extra hybridization steps added for every few cycles to ensure the rest of the transcripts are detected properly. Current technologies include for example the MERFISH method: multiplexed error robust fluorescence in situ hybridization [1].
-> see below the MERFISH technique and the platform based on this technique MERSCOPE from Vizgen.
Vizgen MERSCOPE is a spatial transcriptomics technology that allows for the simultaneous detection of hundreds to thousands of RNA transcripts in tissue samples with subcellular resolution. The MERSCOPE technology is based on a combination of a unique labeling system and imaging technology that enables the detection of RNA transcripts directly in intact tissues. The MERSCOPE workflow involves the hybridization of a set of oligonucleotide probes to RNA transcripts in tissue sections, followed by an enzymatic reaction that generates a series of barcodes specific to each probe-target interaction. These barcodes are then imaged using a specialized microscope that allows for the detection of multiple barcodes simultaneously, providing a highly multiplexed readout of RNA expression. The MERSCOPE technology has the ability to analyze RNA transcripts directly in intact tissue, without the need for tissue dissociation or microdissection. It also provides subcellular resolution and can detect rare cell populations.

[1] Park, J., Kim, J., Lewy, T. et al. Spatial omics technologies at multimodal and single cell/subcellular level. Genome Biol 23, 256 (2022). https://doi.org/10.1186/s13059-022-02824-6
Platforms to contact for this expertise