Chromosome Contact map : 3C, 4C , 5C
It is known that the state of the 3D conformation of chromatin plays a regulatory role on gene expression, replication and DNA repair for a given biological condition of a cell.
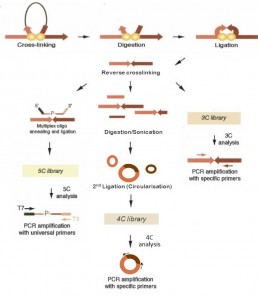
The “capture” of the 3D organization of the chromatin, called “3C”, is a method allowing the identification of chromatin contact points at a region of interest. Chromatin loop conformations and interactions between several regulatory regions were releaved using this approach. Based on the same principle, 4C and 5C techniques have been developed to interrogate networks of connections between very distant regions and to adapt them to high throughput sequencing (Sati & Cavalli 2016).
The 3C-seq method is based on the immobilisation of chromatin conformation and sequencing of interaction regions .
The chromatin is frozen in its 3D conformation with formaldehyde (crosslinking). The DNA is fragmented by enzymatic digestion (HindIII in most cases). The next step is a diluted ligation to promote intramolecular ligation of the restriction fragments. Depending on the approach chosen, the DNA is then sequenced after PCR amplification for the study of interactions at a specific locus or after sonication and size selection for a more global study of chromatin interactions.
Rubriques associées
- Small RNA Sequencing
- Mapping of Transcription Start Sites – TSS
- TAPS/TAPSβ
- Enzymatic Methyl-seq (EM-seq™)
- Methylation of native DNA and RNA
- DNA binding sites map : CUT & RUN vs CUT & Tag
- High Chromosome Contact map : HiC-seq
- Mapping of chromatin accessibility sites: DNase seq
- Indirect mapping of chromatin accessibility sites: MNase seq
- Mapping of chromatin accessibility sites: FAIRE seq
- Mapping of chromatin accessibility sites: ATAC seq
- Mapping of RNA-protein interaction sites: CLIP seq
- Mapping of DNA-protein interaction sites: CHIP seq
- Mapping of DNA epigenetic marks: MeDIP
- Mapping of DNA epigenetic marks: Methyl seq
- BiSeq