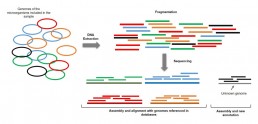
Shotgun metagenomics
This approach provides a global overview of the microbiome composition (bacteria, viruses, fungi, etc…) and its functional properties by identifying the genes that depicted. The consensus sequences obtained after assembling the sequencing reads are aligned with referenced genomes in order to identify the species composing the microbiome. Consensus sequences missing from databases define new species that can in turn be annotated and incorporated into databases (Fig1.)