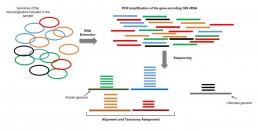
Metagenomics using targeted amplicon/ barcoding/metabarcoding
Genomic studies for the assessment of the composition of a microbiome in terms of species consist in sequencing one or more specific genes called barcodes, shared with several species. For example, the targeted sequencing of the gene encoding 16S rRNA will identify the representation and abundance of the different bacterial species of a given microbiome (Fig1.), or ITS (Internal Transcribed Spacer rRNA) the intraspecific variability in a taxon. The sequencing reads are aligned with the bacterial genomes referenced in databases for identification and classification (taxonomy) of the detected species. Unassigned sequences are ignored. A very wide combination of barecodes can be chosen to detect a wider spectrum of species (metabarcoding).