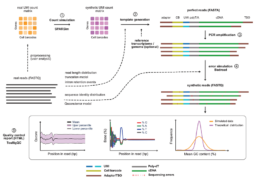
AsaruSim: a single-cell and spatial RNA-Seq Nanopore long-reads simulation workflow
AsaruSim is an open-source workflow developed by the GenomiqueENS genomics platform at IBENS (Institut de Biologie de l’École normale supérieure), which simulates single-cell synthetic long-read Nanopore datasets, accurately reproducing real experimental data. AsaruSim uses a multi-step process that includes the creation of a synthetic count matrix, the generation of perfect reads, optional PCR amplification, the introduction of sequencing errors and a comprehensive quality control report. Applied to a mononuclear blood cell dataset, AsaruSim accurately reproduced experimental read characteristics.
Potential applications include the generation of reads with differential gene expression or DEI between groups of cells, as well as the simulation of known fold changes or batch effects, in order to evaluate and optimise single-cell long-read methods.
Currently, AsaruSim generates data compatible with 10X Genomics 3ʹ and spatial transcriptomics protocols.

AsaruSim source code is available on GitHub :
https://github.com/GenomiqueENS/AsaruSim
Contact : hamraoui@bio.ens.psl.eu