TAPS/TAPSβ
To overcome the biases induced by the BiSeq method for detecting 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) in DNA (DNA degradation, low yield, low complexity, etc.), new methods are emerging for studying epigenetic modifications of DNA.
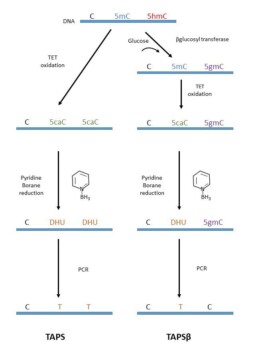
One of these is TAPS (TET-assisted pyridine borane sequencing), which combines oxidation of 5mC and 5hmC by a TET (ten-eleven translocation) enzyme with pyridine borane reduction to generate the C->T transition (see figure).
These reactions induce little DNA damage while maintaining the integrity of fragments up to 10kb, offering the possibility of long-read sequencing (PacBio Chen J et al. N.A.R. 2022; ONT).
If only interested in studying 5mC sites (TAPSβ), it is possible to protect 5hmC by binding a glucose molecule through a β-Glucosyl transferase (Lui Y et al. Nat Comm 2021).
Platforms to contact for this expertise
Rubriques associées
- Small RNA Sequencing
- Mapping of Transcription Start Sites – TSS
- Enzymatic Methyl-seq (EM-seq™)
- Methylation of native DNA and RNA
- DNA binding sites map : CUT & RUN vs CUT & Tag
- Chromosome Contact map : 3C, 4C , 5C
- High Chromosome Contact map : HiC-seq
- Mapping of chromatin accessibility sites: DNase seq
- Indirect mapping of chromatin accessibility sites: MNase seq
- Mapping of chromatin accessibility sites: FAIRE seq
- Mapping of chromatin accessibility sites: ATAC seq
- Mapping of RNA-protein interaction sites: CLIP seq
- Mapping of DNA-protein interaction sites: CHIP seq
- Mapping of DNA epigenetic marks: MeDIP
- Mapping of DNA epigenetic marks: Methyl seq
- BiSeq