Transcriptome-wide or NGS-based approaches
Earlier methods focused on targeted acquisition of sample subsets for sequencing and used laser-capture microscopy (LCM) and photocleavable marker-based methods, where specific regions marked by surface markers or mRNA probes were collected and sequenced. These technologies allow user-directed profiling of specific ROIs with as few as 10 cells, allowing researchers to characterize multiple replicates or tissue types/locations for each sample [1].
–>NanoString GeoMx DSP is a contemporary example of this technology.
Methods that allow single cell or subcellular resolution rely on spatial barcoding and in situ sequencing. These technologies use grid-like nanoballs or sequencing sites on the slide.
–>10X Visium for multi-cell version of the same technology
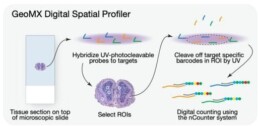
NanoString GeoMx Digital Spatial Profiler (DSP) is a next-generation spatial profiling platform that allows researchers to analyze gene expression, protein expression, and cell morphology in situ, or in their native tissue context. The GeoMx DSP platform is based on a digital barcoding technology that enables high-plex profiling of RNA and protein targets in spatially defined regions of interest within tissue samples [2].
The platform uses a set of DNA barcoded oligonucleotides, called probes, that are designed to target specific RNA or protein molecules of interest. These probes are hybridized to the tissue section, and the resulting hybridization pattern is read out using a digital imaging system. This approach allows for the detection of thousands of RNA or protein targets simultaneously in a single tissue section.

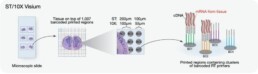
10X Genomics Visium is a spatial transcriptomics technology that allows for the simultaneous analysis of gene expression and spatial information in intact tissue samples. This technology enables researchers to map the transcriptome of individual cells to their specific location within the tissue, providing insights into the spatial organization of gene expression.
The Visium workflow involves the preparation of fresh or fixed tissue samples into thin sections, followed by the permeabilization of the tissue sections and hybridization of oligonucleotide barcoded probes to capture RNA molecules. The captured RNA molecules are then sequenced, and the resulting data are processed using proprietary software to generate spatially resolved gene expression profiles.
One of the key features of the Visium technology is its ability to analyze tissue samples without the need for microdissection or dissociation, which can disrupt the spatial organization of cells and gene expression. This makes it possible to analyze complex tissue structures, such as the brain, where cell-cell interactions and spatial relationships are critical for understanding function and disease.
[1] Park, J., Kim, J., Lewy, T. et al. Spatial omics technologies at multimodal and single cell/subcellular level. Genome Biol 23, 256 (2022). https://doi.org/10.1186/s13059-022-02824-6
[2] Zollinger DR, Lingle SE, Sorg K, Beechem JM, Merritt CR. Geomx™ RNA assay: high multiplex, digital, spatial analysis of RNA in FFPE tissue. Methods Mol Biol. 2020 ; 2148:331–45.

