Spatial transcriptomics
The contribution of single cell analysis is major for biological interpretation. However, tissue organization in space remains an essential information (especially in the study of tumor samples) which is not resolved by conventional single cell techniques.
–> Implementation of Spatial Transcriptomics technologies.
Spatial transcriptomics is a technology that combines high-throughput gene expression profiling with spatial information of cells in tissues or organs. It allows researchers to identify and map the gene expression patterns of individual cells in their native tissue context, providing a more comprehensive understanding of the molecular and cellular organization of tissues.
In traditional transcriptomics, RNA is extracted from a tissue or cell population and sequenced to determine the gene expression profile of the sample. However, this approach does not provide information about the spatial distribution of the expressed genes within the tissue. Spatial transcriptomics addresses this limitation by physically preserving the spatial arrangement of cells within the tissue during the sequencing process.
In scRNA-seq, researchers dissociate cells from tissue to, for example, discern cell types on the basis of gene expression. Now, with spatially resolved transcriptomic methods, scientists can get transcriptomic data and know the positional context of those cells in a tissue [1].
Currently, there are over 50 different spatial mapping technologies available.
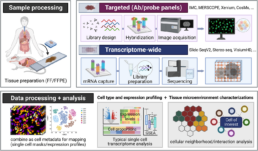
In recent years, traditional experimental methods, including barcoding with reporters, immunohistochemistry (IHC), and fluorescent in situ hybridization (FISH), have given ways to spatial omics technologies to cover a larger number of transcripts or areas (Fig. 1). Broadly, spatial omics technologies vary in their spatial resolution (minimum size of molecular units profiled), coverage (breadth of tissue covered), scale and throughput (number of samples and profiling speed), and multiplexing capacity (breadth of molecular entities profiled simultaneously).
Depending on the research question, the profiling methods can be divided into I) targeted or multiplexed probe- or antibody-based and II) transcriptome-wide or next-generation sequencing (NGS)-based approaches. Most spatial omics technologies with subcellular level resolution are performed on slide (in situ) using either microscopy or NGS platforms [2].
[1] Marx, V. Method of the Year: spatially resolved transcriptomics. Nat Methods 18, 9–14 (2021). https://doi.org/10.1038/s41592-020-01033-y
[2] Park, J., Kim, J., Lewy, T. et al. Spatial omics technologies at multimodal and single cell/subcellular level. Genome Biol 23, 256 (2022). https://doi.org/10.1186/s13059-022-02824-6