scDNA-seq
According to Evrony GD. et al. (2021), single-cell DNA-Seq offers a range of properties that simultaneously address:
– Detection of somatic mutations and CNVs present at a low level of mosaicism
– Evaluation of copresence for lineage definition on a group of differentiated cells
– Detection of DNA features associated with the cell phenotype according to its type and/or biological state
Therefore, the DNA-Seq single cell approach has facilitated for example
– understanding the cellular heterogeneity of cancer tumours and their evolution
– the identification of the composition of an ecosystem microbiota and the resulting phylogenetic relationships
– monitoring pathogens in an ecosystem
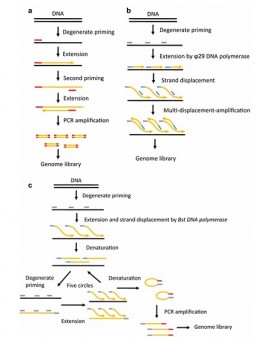
An important limitation of such application resides in the fact that the amount of DNA per cell is too small to be used on the commercially available sequencers. This means that a genomic amplification step (WGA) must be included in the library preparation protocol, with the risk of introducing errors that could disrupt the bioinformatics analyses. Several methods (see figure) can be used, each with their pros and cons (Wang J and Song Y 2017)
– DOP-PCR (a): PCR with degenerate primers to detect CNVs but not SNPs
– MDA (b): displacement amplification to detect SNPs but with heterogeneous genome coverage and chimeric sequences that pollute the analyses
– MALBAC (c): pre-displacement amplification followed by loop PCR allowing good amplification fidelity and homogeneity across the whole genome for the detection of CNVs and SNPs.
To apply WGA at high throughput Fu Y. et al. (2019) suggests the MICA-eMDA protocol which introduces the generation of an emulsion where each droplet hosts a pre amplification.
Platforms to contact for this expertise