DNA binding sites map : CUT & RUN vs CUT & Tag
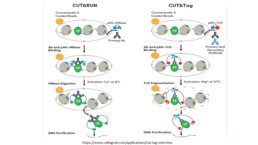
CUT & RUN (Cleavage Under Target & Release Using Nuclease) is part of the series of protocols (CHIP, ATAC, FAIRE seq….) used to map nucleosomes and chromatin accessibility for transcription regulatory factors. This new method has the advantage of operating in situ, on intact cells or nuclei, avoiding the crosslinking and fragmentation steps that produce DNA fragments outside / far away from the DNA-protein interaction sites generating to a lot of background sequencing noise. Depending on the specificity of the antibody, CUT & RUN allows targeting histone modifications, transcriptional factors and co-factors (PJ Sken and S Henikoff, 2017).
The CUT & RUN method essentially consists in immobilizing permeabilized cells or nuclei on magnetic beads and incubating them with antibodies specific to the target proteins (histone modifications, transcription factors, etc.) and then with Protein A-MNase, both diffusing through the pores of the nucleus. Once the target-antibody – Protein A-MNase binding is completed, Ca++ is added at low temperature to activate Protein A-MNase and cleave the DNA at both ends of the DNA-target protein complex. The fragments are recovered and the DNA is purified to go through the library preparation protocol for sequencing. Approximately 3 to 5M reads are sufficient for analysis (about 1/10 of the coverage required for CHIP seq).
The performance of the method depends on the specificity of the antibody and the abundance of the target.
This protocol has been adapted for studies on single cells and nuclei using CUT & Tag (Kaya-Okur, H.S. et al. 2020), which uses pA-Tn5 that can be indexed with adapters. Its activation in the presence of Mg++ leads to tagmentation with integration of the adapters into the target DNA, thus avoiding the ligation step in the preparation of the library for sequencing (see figure).
Platforms to contact for “CUT & RUN” sequencing projects
Last update April 2024
Rubriques associées
- Small RNA Sequencing
- Mapping of Transcription Start Sites – TSS
- TAPS/TAPSβ
- Enzymatic Methyl-seq (EM-seq™)
- Methylation of native DNA and RNA
- High Chromosome Contact map : HiC-seq
- Indirect mapping of chromatin accessibility sites: MNase seq
- Mapping of chromatin accessibility sites: ATAC seq
- Mapping of RNA-protein interaction sites: CLIP seq
- Mapping of DNA-protein interaction sites: CHIP seq
- Mapping of DNA epigenetic marks: MeDIP
- Mapping of DNA epigenetic marks: Methyl seq
- BiSeq