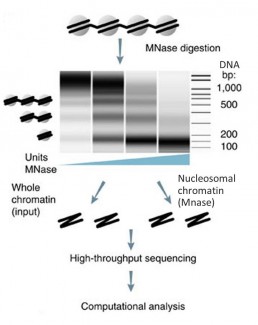
Indirect mapping of chromatin accessibility sites: MNase seq
Nucleosome arrangement is an important aspect of chromatin organisation, with direct implications on gene regulation. The usual nucleosome mapping technique is based on micrococcal nuclease (MNase) digestion, which preferentially targets free DNA (unprotected by proteins). When compared to a control sample that has not been MNAse-digested, chromatin accessibility sites can be indirectly determined.
To avoid any re-conformation of the chromatin during its extraction or fragmentation, crosslinking is recommended using formaldehyde. The MNase enzymatic digestion is a critical step in the protocol, as concentrations and incubation times should be chosen so that all classes of nucleosomes can be isolated and mononucleosomal DNA fragments of 200bp to 500bp can be obtained without leading to DNA damage (see figure). . The DNA-histone complexes are then cleaved and the purified DNA can be processed for sequencing (Mieczkowski et al. 2016).
Rubriques associées
- Small RNA Sequencing
- Mapping of Transcription Start Sites – TSS
- TAPS/TAPSβ
- Enzymatic Methyl-seq (EM-seq™)
- Methylation of native DNA and RNA
- DNA binding sites map : CUT & RUN vs CUT & Tag
- Chromosome Contact map : 3C, 4C , 5C
- High Chromosome Contact map : HiC-seq
- Mapping of chromatin accessibility sites: DNase seq
- Mapping of chromatin accessibility sites: FAIRE seq
- Mapping of chromatin accessibility sites: ATAC seq
- Mapping of RNA-protein interaction sites: CLIP seq
- Mapping of DNA-protein interaction sites: CHIP seq
- Mapping of DNA epigenetic marks: MeDIP
- Mapping of DNA epigenetic marks: Methyl seq
- BiSeq