Small RNA Sequencing
Non-coding small RNAs (ncRNA, smallRNA) and microRNAs (miRNA) are key regulators of gene expression, heterochromatin formation and nucleus organisation. Their actions are dependent on tissue and environment and, through various mechanisms, they have a major influence on cancer and numerous diseases.
Small non-coding RNAs (ncRNA, smallRNA) and microRNAs (miRNA) are major regulators of gene expression, heterochromatin formation and nucleus organization. Their action is dependent on tissue, the environment, and through various mechanisms, has a major influence in cancers and many diseases.
The extraction of small RNAs is a tricky step. Several methods are available: from the total RNAs, the small RNAs will be separated by gel electrophoresis, elution through a silica membrane or purification on magnetic beads (Wong et al.).
Preparing libraries for NGS sequencing is not without its challenges, as it involves a high risk of bias in the representation of small RNAs and contamination by adaptor dimers (Coenenen-Stass et al.).
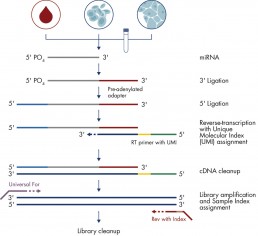
In brief, the protocol includes the following steps: addition of a specific adaptor at each end of the RNA molecules, conversion of the RNA into double-stranded cDNA (reverse transcription) including, depending on the kit used, molecule indexing (UMI) and finally amplification. The libraries are then quality controlled on Bioanalyzer (Agilent) and quantified with QuBit (Invitrogen).
Rubriques associées
- Mapping of Transcription Start Sites – TSS
- TAPS/TAPSβ
- Enzymatic Methyl-seq (EM-seq™)
- Methylation of native DNA and RNA
- DNA binding sites map : CUT & RUN vs CUT & Tag
- Chromosome Contact map : 3C, 4C , 5C
- High Chromosome Contact map : HiC-seq
- Mapping of chromatin accessibility sites: DNase seq
- Indirect mapping of chromatin accessibility sites: MNase seq
- Mapping of chromatin accessibility sites: FAIRE seq
- Mapping of chromatin accessibility sites: ATAC seq
- Mapping of RNA-protein interaction sites: CLIP seq
- Mapping of DNA-protein interaction sites: CHIP seq
- Mapping of DNA epigenetic marks: MeDIP
- Mapping of DNA epigenetic marks: Methyl seq
- BiSeq