Genotyping (DNA, RNA, CpG arrays)
Unlike DNA re-sequencing, which allows the blind screening of genome variants (SNP polymorphisms, indels, LOH…), genotyping aims to identify changes at known positions, on part or all over of the genome.
Depending on the objective of the genotyping project, several technologies are available.
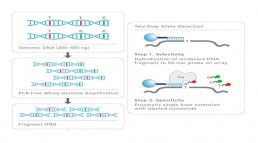
Genome Wide Association Study (GWAS) and replication studies require questioning the polymorphism of several million variants distributed over the entire length of the genome on large cohorts. Very high throughput technologies (Illumina, Agilent, Affymetrix…), based on the hybridization of DNA to oligonucleotides that are specific to genetic variations, are used for this type of project (Fig1).
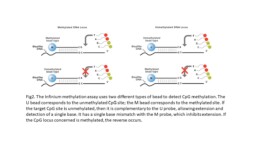
In addition to sequence polymorphisms, the same approach can be used to determine the degree of methylation of CpG islands. The DNA is first treated with bisulphite, which converts unmethylated cytosines to uracil. The level of methylation is determined by the ratio between the signals from methylated sites and unmethylated sites (Fig2).
The Sequenom spectrometer using MALDI-TOF technology, Matrix-Assisted Laser Desorption/Ionisation Time of Flight, is suitable for replication/ validation studies of candidate regions from a GWAS (several dozen variants).
Applied Biosystems’ 7900-HT system, using Taqman chemistry based on allelic discrimination by amplification, is used for fine mapping (genotyping of a few variants).