The sequencing of the Regulome
The regulome refers to the interaction between all the regulatory components of a cell that control the state of expression of genes and their isoforms, according to their subcellular location, tissue state, development and pathology.
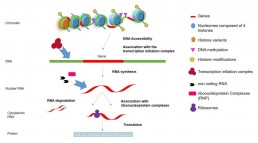
Regulation can occur at several stages (Fig1.).
1- The accessibility of DNA for gene transcription depends on factors that can modify the compaction of the 5 families of histones on which the DNA is wrapped (Tsompana & Buck 2014; Klein & Hainer 2020; Sati &Cavalli 2016).).
2- The level of DNA methylation (addition/deletion of CH3 groups on Cytosine) modulates gene transcription.
3- Regulatory proteins (transcription factors) have the ability to bind specifically to short DNA sequences and to regulate the transcription of a gene in a positive or negative way.
4- RNA-protein interactions (RNP: ribonucleoprotein complexes) act on post-trancriptional mechanisms such as splicing, RNA stability, etc.
5- Different classes of non-coding RNAs such as small and micro RNAs can be involved in splicing, ribosome synthesis, degradation of mRNAs, translation repression.
All these mechanisms are affected by environmental factors (diet, pollution, age, stress, medication, etc.)
Rubriques associées
- Small RNA Sequencing
- Mapping of Transcription Start Sites – TSS
- TAPS/TAPSβ
- Enzymatic Methyl-seq (EM-seq™)
- Methylation of native DNA and RNA
- DNA binding sites map : CUT & RUN vs CUT & Tag
- Chromosome Contact map : 3C, 4C , 5C
- High Chromosome Contact map : HiC-seq
- Mapping of chromatin accessibility sites: DNase seq
- Indirect mapping of chromatin accessibility sites: MNase seq
- Mapping of chromatin accessibility sites: FAIRE seq
- Mapping of chromatin accessibility sites: ATAC seq
- Mapping of RNA-protein interaction sites: CLIP seq
- Mapping of DNA-protein interaction sites: CHIP seq
- Mapping of DNA epigenetic marks: MeDIP
- Mapping of DNA epigenetic marks: Methyl seq
- BiSeq