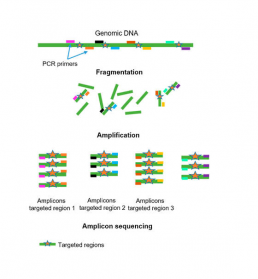
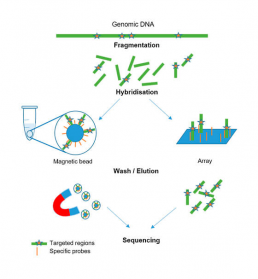
The squencing of the targeted genome
Targeted re-sequencing allows efficient identification of variants (SNPs, deletions, insertions) that may appear in regions of interest. This technique is particularly used in the study of a panel of genes associated with a pathology in a population, or family of genes for the determination of phylogenetic trees. There are two approaches to this sequencing:
– sequencing DNA fragments of interest amplified by PCR (amplicon) (Fig1).
– sequencing DNA fragments of interest “captured” by hybridization to specific probes in the genetic regions to be studied (bait). Two different protocols can be used: solid or in solution capture. In the case of solid capture, the probes are fixed on a solid support (chip/array), while in solution capture, they are attached to magnetic beads (Fig2).